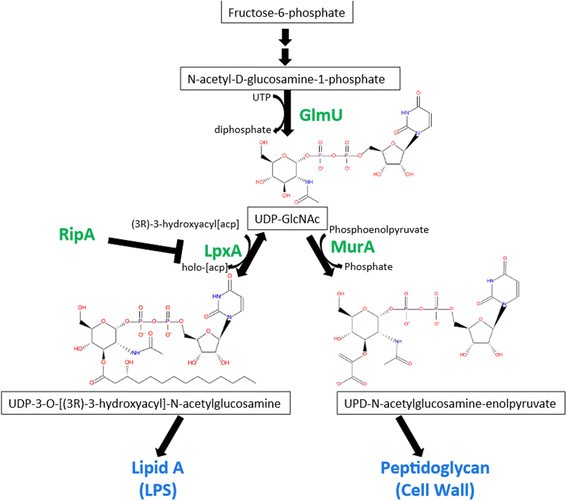
Extragenic suppressor mutations in ΔripA disrupt stability and function of LpxA | BMC Microbiology | Full Text

Evaluation of UDP‐GlcN Derivatives for Selective Labeling of 5‐(Hydroxymethyl)cytosine - Dai - 2013 - ChemBioChem - Wiley Online Library

Structure of UDP-N-acetylglucosamine acyltransferase with a bound antibacterial pentadecapeptide | PNAS

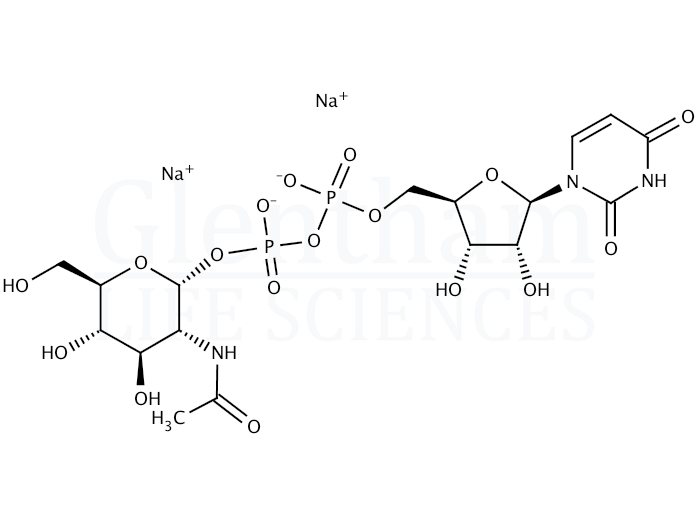
The synthesis of Uridine diphospho--N-acetylglucosamine (UDP-GlcNAc)... | Download Scientific Diagram
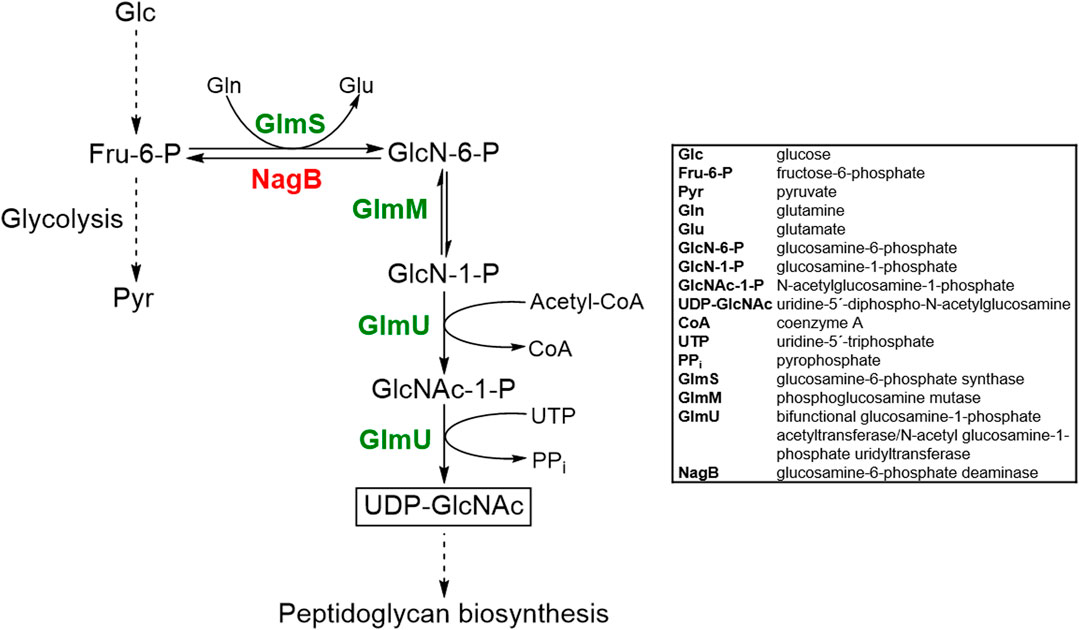
Frontiers | Metabolic Engineering of Corynebacterium glutamicum for Production of UDP-N-Acetylglucosamine

Artificial N-functionalized UDP-glucosamine analogues as modified substrates for N-acetylglucosaminyl transferases - ScienceDirect

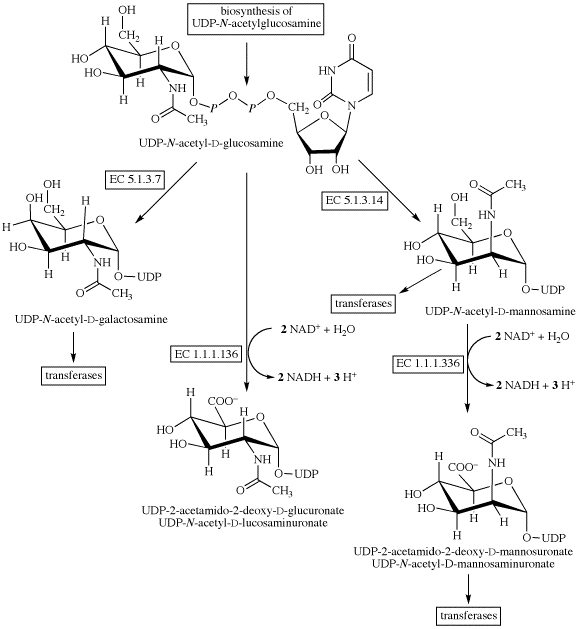
UDP-GlcNAc biosynthetic pathway. The four main enzymes of the pathway... | Download Scientific Diagram
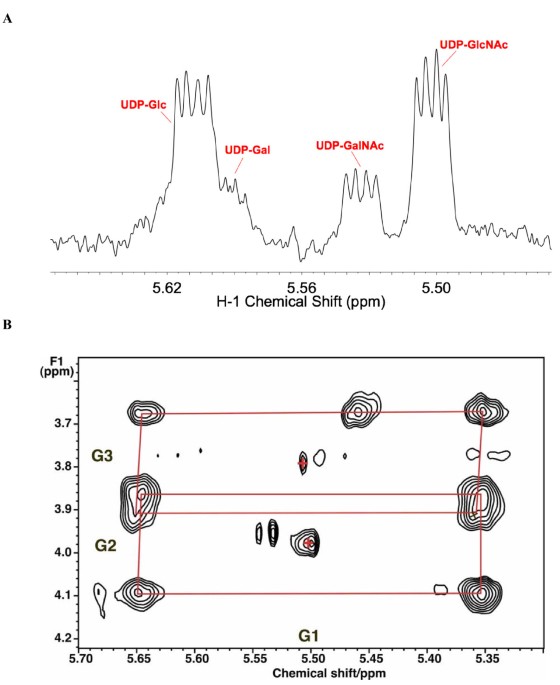
A novel deconvolution method for modeling UDP-N-acetyl-D-glucosamine biosynthetic pathways based on 13C mass isotopologue profiles under non-steady-state conditions | BMC Biology | Full Text
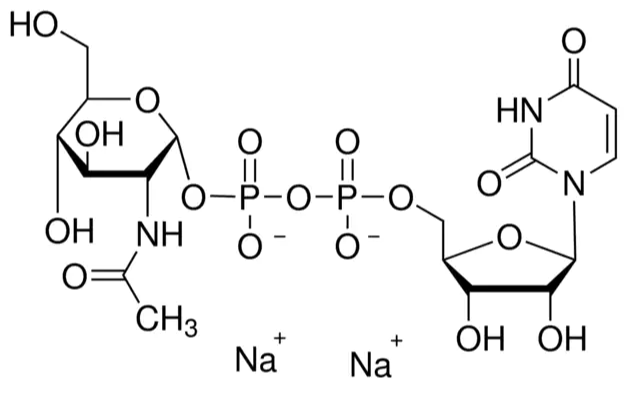
Ligand view of UDP-N-acetyl-alpha-D-glucosamine (112022 - LFTYTUAZOPRMMI-CFRASDGPSA-N) - BRENDA Enzyme Database

A novel deconvolution method for modeling UDP-N-acetyl-D-glucosamine biosynthetic pathways based on 13C mass isotopologue profiles under non-steady-state conditions – topic of research paper in Biological sciences. Download scholarly article PDF and read
BioChemSyn_Sugar Nucleotide_Rare sugar nucleotide_unnatural sugar nucleotide_Rare sugar_UDP-GalNAz, UDP-GlcNAz, GDP-Fucose,CMP-Neu5Ac,HMO,UDP -Gal,UDP-GalNAc,GDP-FucAz, UDP-GlcA,UDP-Glucoronic Acid
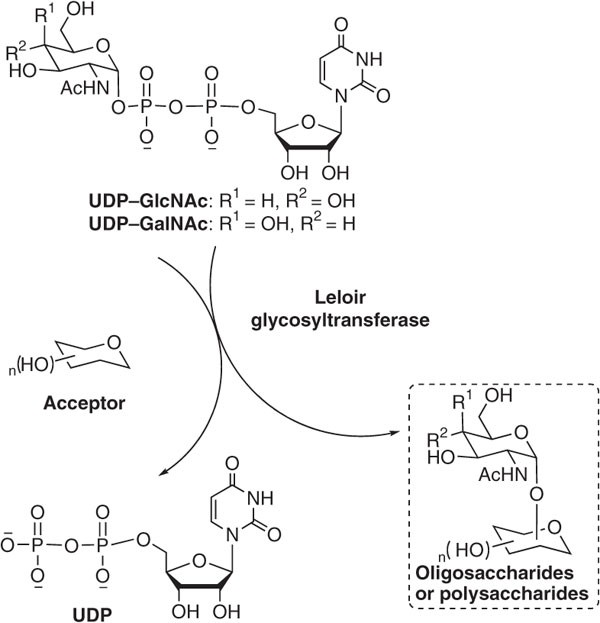
Enzymatic route to preparative-scale synthesis of UDP–GlcNAc/GalNAc, their analogues and GDP–fucose | Nature Protocols

Conversion of UDP-N-glucosamine to UDP-N-acetylmuramyl pentapeptide by... | Download Scientific Diagram
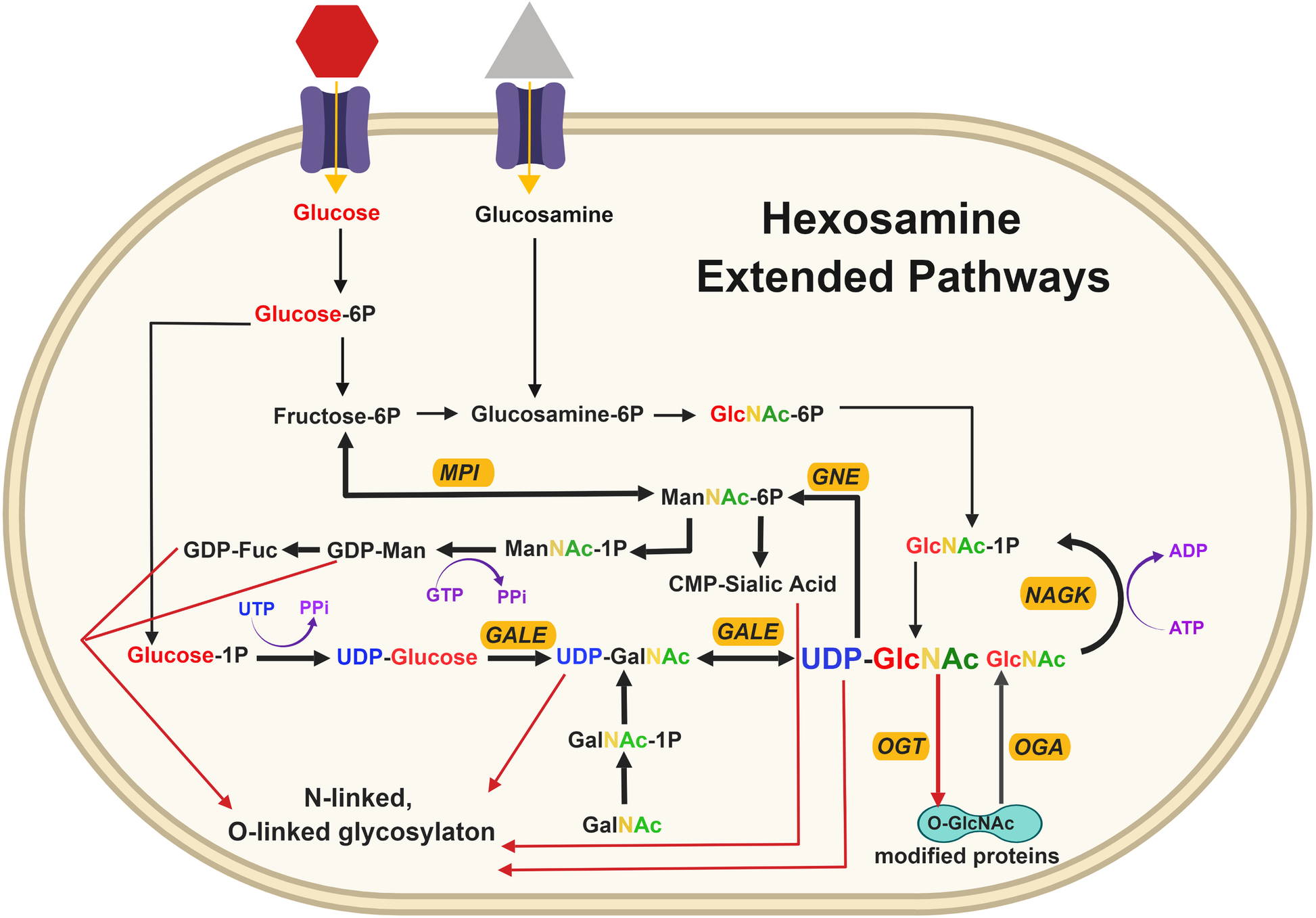
Fueling the fire: emerging role of the hexosamine biosynthetic pathway in cancer | BMC Biology | Full Text
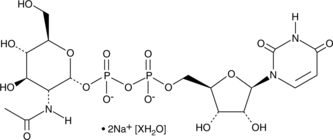
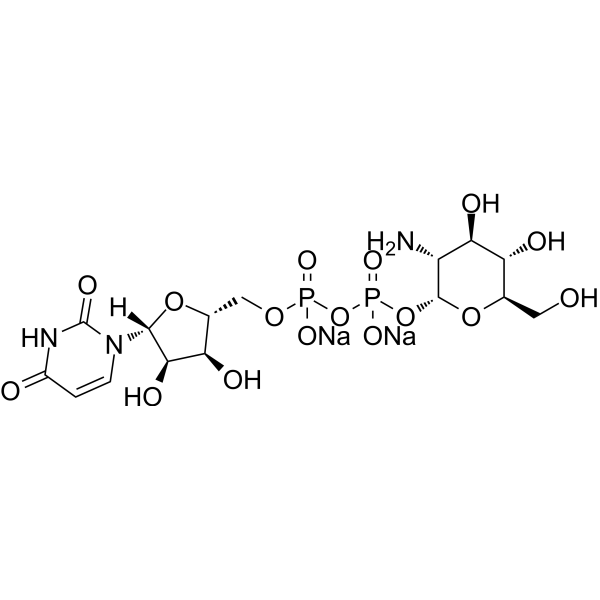

![uridine diphosphate-alpha-N-acetyl-D-[UL-13C6]glucosamine Carbon-13 isotope structure, pricing uridine diphosphate-alpha-N-acetyl-D-[UL-13C6]glucosamine Carbon-13 isotope structure, pricing](https://www.omicronbio.com/pngs/NTS-009.png)